Synaptic Connectivity
Contents
BrainPy provides several commonly used connection methods in brainpy.connect module (see the follows). They are all inherited from the base class brainpy.connect.Connector. Users can also customize their synaptic connectivity by the class inheritance.
[1]:
import brainpy as bp
import numpy as np
import matplotlib.pyplot as plt
Build-in regular connections
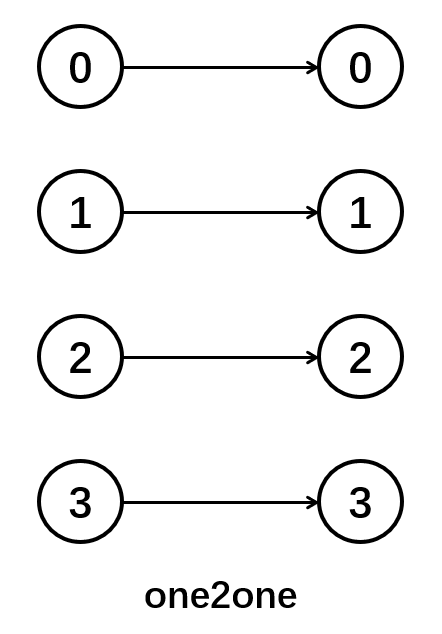
brainpy.connect.One2One
The neurons in the pre-synaptic neuron group only connect to the neurons in the same position of the post-synaptic group. Thus, this connection requires the indices of two neuron groups same. Otherwise, an error will occurs.
[2]:
conn = bp.connect.One2One()
brainpy.connect.All2All
All neurons of the post-synaptic population form connections with all neurons of the pre-synaptic population (dense connectivity). Users can choose whether connect the neurons at the same position (include_self=True or False).

[3]:
conn = bp.connect.All2All(include_self=False)
brainpy.connect.GridFour
GridFour is the four nearest neighbors connection. Each neuron connect to its nearest four neurons.

[4]:
conn = bp.connect.GridFour(include_self=False)
brainpy.connect.GridEight
GridEight is eight nearest neighbors connection. Each neuron connect to its nearest eight neurons.

[5]:
conn = bp.connect.GridEight(include_self=False)
brainpy.connect.GridN
GridN is also a nearest neighbors connection. Each neuron connect to its nearest \(2N \cdot 2N\) neurons.

[6]:
conn = bp.connect.GridN(N=2, include_self=False)
Build-in random connections
brainpy.connect.FixedProb
For each post-synaptic neuron, there is a fixed probability that it forms a connection with a neuron of the pre-synaptic population. It is basically a all_to_all projection, except some synapses are not created, making the projection sparser.

[7]:
conn = bp.connect.FixedProb(prob=0.5, include_self=False, seed=1234)
brainpy.connect.FixedPreNum
Each neuron in the post-synaptic population receives connections from a fixed number of neurons of the pre-synaptic population chosen randomly. It may happen that two post-synaptic neurons are connected to the same pre-synaptic neuron and that some pre-synaptic neurons are connected to nothing.

[8]:
conn = bp.connect.FixedPreNum(num=10, include_self=True, seed=1234)
brainpy.connect.FixedPostNum
Each neuron in the pre-synaptic population sends a connection to a fixed number of neurons of the post-synaptic population chosen randomly. It may happen that two pre-synaptic neurons are connected to the same post-synaptic neuron and that some post-synaptic neurons receive no connection at all.

[9]:
conn = bp.connect.FixedPostNum(num=10, include_self=True, seed=1234)
brainpy.connect.GaussianProb
Builds a Gaussian connection pattern between the two populations, where the connection probability decay according to the gaussian function.
Specifically,
where \((x, y)\) is the position of the pre-synaptic neuron and \((x_c,y_c)\) is the position of the post-synaptic neuron.
For example, in a \(30 \textrm{x} 30\) two-dimensional networks, when \(\beta = \frac{1}{2\sigma^2} = 0.1\), the connection pattern is shown as the follows:

[10]:
conn = bp.connect.GaussianProb(sigma=0.2, p_min=0.01, normalize=True, include_self=True, seed=1234)
brainpy.connect.GaussianWeight
Builds a Gaussian connection pattern between the two populations, where the weights decay with gaussian function.
Specifically,
where \((x, y)\) is the position of the pre-synaptic neuron (normalized to [0,1]) and \((x_c,y_c)\) is the position of the post-synaptic neuron (normalized to [0,1]), \(w_{max}\) is the maximum weight. In order to void creating useless synapses, \(w_{min}\) can be set to restrict the creation of synapses to the cases where the value of the weight would be superior to \(w_{min}\). Default is \(0.01 w_{max}\).
[11]:
def show_weight(pre_ids, post_ids, weights, geometry, neu_id):
height, width = geometry
ids = np.where(pre_ids == neu_id)[0]
post_ids = post_ids[ids]
weights = weights[ids]
X, Y = np.arange(height), np.arange(width)
X, Y = np.meshgrid(X, Y)
Z = np.zeros(geometry)
for id_, weight in zip(post_ids, weights):
h, w = id_ // width, id_ % width
Z[h, w] = weight
fig = plt.figure()
ax = fig.gca(projection='3d')
surf = ax.plot_surface(X, Y, Z, cmap=plt.cm.coolwarm, linewidth=0, antialiased=False)
fig.colorbar(surf, shrink=0.5, aspect=5)
plt.show()
[12]:
conn = bp.connect.GaussianWeight(sigma=0.1, w_max=1., w_min=0.01,
normalize=True, include_self=True)
[13]:
pre_geom = post_geom = (40, 40)
conn(pre_geom, post_geom)
pre_ids = conn.pre_ids
post_ids = conn.post_ids
weights = conn.weights
show_weight(pre_ids, post_ids, weights, pre_geom, 820)

brainpy.connect.DOG
Builds a Difference-Of-Gaussian (dog) connection pattern between the two populations.
Mathematically,
where weights smaller than \(0.01 * abs(w_{max} - w_{min})\) are not created and self-connections are avoided by default (parameter allow_self_connections).
[14]:
dog = bp.connect.DOG(sigmas=(0.08, 0.15), ws_max=(1.0, 0.7), w_min=0.01,
normalize=True, include_self=True)
h = 40
pre_geom = post_geom = (h, h)
dog(pre_geom, post_geom)
pre_ids = dog.pre_ids
post_ids = dog.post_ids
weights = dog.weights
show_weight(pre_ids, post_ids, weights, (h, h), h * h // 2 + h // 2)

brainpy.connect.SmallWorld
SmallWorld is a connector class to help build a small-world network [1]. small-world network is defined to be a network where the typical distance L between two randomly chosen nodes (the number of steps required) grows proportionally to the logarithm of the number of nodes N in the network, that is:
[1] Duncan J. Watts and Steven H. Strogatz, Collective dynamics of small-world networks, Nature, 393, pp. 440–442, 1998.
Currently, SmallWorld only support a one-dimensional network with the ring structure. It receives four settings:
num_neighbor: the number of the nearest neighbors to connect.prob: the probability of rewiring each edge.directed: whether the edge is the directed (“directed=True”) or undirected (“directed=False”) connection.include_self: whether allow to connect to itself.
[15]:
conn = bp.connect.SmallWorld(num_neighbor=5, prob=0.2, directed=False, include_self=False)
brainpy.connect.ScaleFreeBA
ScaleFreeBA is a connector class to help build a random scale-free network according to the Barabási–Albert preferential attachment model [2]. ScaleFreeBA receives the following settings:
m: Number of edges to attach from a new node to existing nodes.directed: whether the edge is the directed (“directed=True”) or undirected (“directed=False”) connection.seed: Indicator of random number generation state.
[2] A. L. Barabási and R. Albert “Emergence of scaling in random networks”, Science 286, pp 509-512, 1999.
[16]:
conn = bp.connect.ScaleFreeBA(m=5, directed=False, seed=12345)
brainpy.connect.ScaleFreeBADual
ScaleFreeBADual is a connector class to help build a random scale-free network according to the dual Barabási–Albert preferential attachment model [3]. ScaleFreeBA receives the following settings:
p: The probability of attaching \(m_1\) edges (as opposed to \(m_2\) edges).m1: Number of edges to attach from a new node to existing nodes with probability \(p\).m2: Number of edges to attach from a new node to existing nodes with probability \(1-p\).directed: whether the edge is the directed (“directed=True”) or undirected (“directed=False”) connection.seed: Indicator of random number generation state.
[3] N. Moshiri. “The dual-Barabasi-Albert model”, arXiv:1810.10538.
[17]:
conn = bp.connect.ScaleFreeBADual(m1=3, m2=5, p=0.5, directed=False, seed=12345)
brainpy.connect.PowerLaw
PowerLaw is a connector class to help build a random graph with powerlaw degree distribution and approximate average clustering [4]. It receives the following settings:
m: the number of random edges to add for each new nodep: Probability of adding a triangle after adding a random edgedirected: whether the edge is the directed (“directed=True”) or undirected (“directed=False”) connection.seed: Indicator of random number generation state.
[4] P. Holme and B. J. Kim, “Growing scale-free networks with tunable clustering”, Phys. Rev. E, 65, 026107, 2002.
[18]:
conn = bp.connect.PowerLaw(m=3, p=0.5, directed=False, seed=12345)
Customize your connections
BrainPy also allows you to customize your model connections. What need users do is only two aspects:
Your connection class should inherit
brainpy.connect.Connector.Initialize the
conn_matorpre_ids+post_idssynaptic structures.Provide
num_preandnum_postinformation.
In such a way, based on this customized connection class, users can generate any other synaptic structures (such like pre2post, pre2syn, pre_slice_syn, etc.) easily.
Here, let’s take a simple connection as an example. In this example, we create a connection method which receives users’ handful index projection.
[19]:
class IndexConn(bp.connect.Connector):
def __init__(self, i, j):
super(IndexConn, self).__init__()
# initialize the class via "pre_ids" and "post_ids"
self.pre_ids = bp.ops.as_tensor(i)
self.post_ids = bp.ops.as_tensor(j)
def __call__(self, pre_size, post_size):
self.num_pre = bp.size2len(pre_size) # this is ncessary when create "pre2post" ,
# "pre2syn" etc. structures
self.num_post = bp.size2len(post_size) # this is ncessary when create "post2pre" ,
# "post2syn" etc. structures
return self
Let’s try to use it.
[20]:
conn = IndexConn(i=[0, 1, 2], j=[0, 0, 0])
conn = conn(pre_size=5, post_size=3)
[21]:
conn.requires('conn_mat')
[21]:
array([[1., 0., 0.],
[1., 0., 0.],
[1., 0., 0.],
[0., 0., 0.],
[0., 0., 0.]])
[22]:
conn.requires('pre2post')
[22]:
[array([0]),
array([0]),
array([0]),
array([], dtype=int32),
array([], dtype=int32)]
[23]:
conn.requires('pre2syn')
[23]:
[array([0]),
array([1]),
array([2]),
array([], dtype=int32),
array([], dtype=int32)]
[24]:
conn.requires('pre_slice_syn')
[24]:
array([[0, 1],
[1, 2],
[2, 3],
[3, 3],
[3, 3]])
Author:
Chaoming Wang
Email: adaduo@outlook.com
Date: 2021.04.16