Customizing Your Synapse Models#
Synaptic computation is the core of brain dynamics programming. This is because in a real project most of the simulation time spends on the computation of synapses. In order to achieve efficient synaptic computation, BrainPy provides many useful supports. Here, we are going to explore the details of these supports.
import brainpy as bp
import brainpy.math as bm
bm.set_platform('cpu')
bp.__version__
'2.4.0'
Synapse Models in Math#
Before we talk about the implementation of synapses in BrainPy, it’s better to understand the targets (synapse models) we are going to implement. For different illustration purposes, we are going to implement two synapse models: exponential synapse model and AMPA synapse model.
1. The exponential synapse model#
The exponential synapse model assumes that once a pre-synaptic neuron generates a spike, the synaptic state arises instantaneously, then decays with a certain time constant \(\tau_{decay}\). Its dynamics is given by:
where \(g\) is the synaptic state, \(t^{k}\) is the spike time of the pre-synaptic neuron, and \(D\) is the synaptic delay.
Afterward, the current output onto the post-synaptic neuron is given in the conductance-based form:
where \(E\) is the reversal potential of the synapse, \(V\) is the post-synaptic membrane potential, \(g_{max}\) is the maximum synaptic conductance.
2. The AMPA synapse model#
A classical model of AMPA synapse is to use the Markov process to model ion channel switch. Here \(g\) represents the probability of channel opening, \(1-g\) represents the probability of ion channel closing, and \(\alpha\) and \(\beta\) are the transition probability. Specifically, its formula is given by
where \(\alpha [T]\) denotes the transition probability from state \((1-g)\) to state \((g)\); and \(\beta\) represents the transition probability of the other direction. \(\alpha\) is the binding constant. \(\beta\) is the unbinding constant. \([T]\) is the neurotransmitter concentration, and has the duration of 0.5 ms.
Moreover, the post-synaptic current on the post-synaptic neuron is formulated as
where \(g_{max}\) is the maximum conductance, and \(E\) is the reverse potential.
Synapse Models in Silicon#
The implementation of synapse models is accomplished by brainpy.TwoEndConn interface. In this section, we talk about what supports are provided for the implementation of synapse models in silicon.
1. brainpy.SynConn#
In BrainPy, brainpy.SynConn is used to model two-end synaptic computations.
To define a synapse model, two requirements should be satisfied:
1. Constructor function __init__(), in which three key arguments are needed.
pre: the pre-synaptic neural group. It should be an instance ofbrainpy.NeuGroup.post: the post-synaptic neural group. It should be an instance ofbrainpy.NeuGroup.conn(optional): the connection type between these two groups. BrainPy has provided abundant connection types that are described in details in the Synaptic Connections.
2. Update function update(tdi) describes the updating rule from the current time \(\mathrm{tdi.t}\) to the next time \(\mathrm{tdi.t + tdi.dt}\).
2. Variable delays#
As seen in the above two synapse models, synaptic computations are usually involved with variable delays. A delay time (typically 0.3–0.5 ms) is usually required for a neurotransmitter to be released from a pre-synaptic membrane, diffuse across the synaptic cleft, and bind to a receptor site on the post-synaptic membrane.
BrainPy provides several kinds of delay variables for users, including:
brainpy.math.LengthDelay: a delay variable which defines a constant steps for delay.brainpy.math.TimeDelay: a delay variable which defines a constant time length for delay.
Assume here we need a delay variable which has 1 ms delay. If the numerical integration precision dt is 0.1 ms, then we can create a brainpy.math.LengthDelay which has 10 delay time steps.
target_data_to_delay = bm.Variable(bm.zeros(10))
example_delay = bm.LengthDelay(target_data_to_delay,
delay_len=10) # delay 10 steps
example_delay(5) # call the delay data at 5 delay step
DeviceArray([0., 0., 0., 0., 0., 0., 0., 0., 0., 0.], dtype=float32)
example_delay(10) # call the delay data at 10 delay step
DeviceArray([0., 0., 0., 0., 0., 0., 0., 0., 0., 0.], dtype=float32)
Alternatively, we can create an instance of brainpy.math.TimeDelay, which use time t as the index to retrieve the delay data.
t0 = 0.
example_delay = bm.TimeDelay(target_data_to_delay,
delay_len=1.0, t0=t0) # delay 1.0 ms
example_delay(t0 - 1.0) # the delay data at t-1. ms
DeviceArray([0., 0., 0., 0., 0., 0., 0., 0., 0., 0.], dtype=float32)
example_delay(t0 - 0.5) # the delay data at t-0.5 ms
DeviceArray([0., 0., 0., 0., 0., 0., 0., 0., 0., 0.], dtype=float32)
3. Synaptic connections#
Synaptic computations usually need to create connection between groups. BrainPy provides many wonderful supports to construct synaptic connections. Simply speaking, brainpy.conn.Connector can create various data structures you want through the require() function. Take the random connection brainpy.conn.FixedProb which will be used in follows as the example,
example_conn = bp.conn.FixedProb(0.2)(pre_size=(5,), post_size=(8, ))
we can require the connection matrix (has the shape of (num_pre, num_post):
example_conn.require('conn_mat')
DeviceArray([[ True, False, False, False, False, True, False, True],
[False, False, False, False, False, False, False, False],
[False, False, False, False, True, True, False, True],
[False, False, False, False, False, False, False, False],
[False, True, False, False, True, True, False, False]], dtype=bool)
we can also require the connected indices of pre-synaptic neurons (pre_ids) and post-synaptic neurons (post_ids):
example_conn.require('pre_ids', 'post_ids')
(DeviceArray([0, 1, 2, 3, 4], dtype=int32),
DeviceArray([2, 7, 4, 2, 1], dtype=int32))
Or, we can require the connection structure of pre2post which stores the information how does each pre-synaptic neuron connect to post-synaptic neurons:
example_conn.require('pre2post')
(DeviceArray([7, 2, 3, 5, 4], dtype=int32),
DeviceArray([0, 1, 2, 3, 4, 5], dtype=int32))
Warning
Every require() function will establish a new connection pattern, and return the data structure users have required. Therefore any two require() will return different connection pattern, just like the examples above. Please keep in mind to require all the data structure at once if users want a consistent connection pattern.
More details of the connection structures please see the tutorial of Synaptic Connections.
Achieving efficient synaptic computation is difficult#
Synaptic computations usually need to transform the data of the pre-synaptic dimension into the data of the post-synaptic dimension, or the data with the shape of the synapse number. There does not exist a universal computation method that are efficient in all cases. Usually, we need different ways for different connection situations to achieve efficient synaptic computation. In the next two sections, we will talk about how to define efficient synaptic models when your connections are sparse or dense.
Before we start, we need to define some useful helper functions to define and show synapse models. Then, we will highlight the key differences of model definition when using different synaptic connections.
# Basic Model to define the exponential synapse model. This class
# defines the basic parameters, variables, and integral functions.
class BaseExpSyn(bp.SynConn):
def __init__(self, pre, post, conn, g_max=1., delay=0., tau=8.0, E=0., method='exp_auto'):
super(BaseExpSyn, self).__init__(pre=pre, post=post, conn=conn)
# check whether the pre group has the needed attribute: "spike"
self.check_pre_attrs('spike')
# check whether the post group has the needed attribute: "input" and "V"
self.check_post_attrs('input', 'V')
# parameters
self.E = E
self.tau = tau
self.delay = delay
self.g_max = g_max
# use "LengthDelay" to store the spikes of the pre-synaptic neuron group
self.delay_step = int(delay/bm.get_dt())
self.pre_spike = bm.LengthDelay(pre.spike, self.delay_step)
# integral function
self.integral = bp.odeint(lambda g, t: -g / self.tau, method=method)
# Basic Model to define the AMPA synapse model. This class
# defines the basic parameters, variables, and integral functions.
class BaseAMPASyn(bp.SynConn):
def __init__(self, pre, post, conn, delay=0., g_max=0.42, E=0., alpha=0.98,
beta=0.18, T=0.5, T_duration=0.5, method='exp_auto'):
super(BaseAMPASyn, self).__init__(pre=pre, post=post, conn=conn)
# check whether the pre group has the needed attribute: "spike"
self.check_pre_attrs('spike')
# check whether the post group has the needed attribute: "input" and "V"
self.check_post_attrs('input', 'V')
# parameters
self.delay = delay
self.g_max = g_max
self.E = E
self.alpha = alpha
self.beta = beta
self.T = T
self.T_duration = T_duration
# use "LengthDelay" to store the spikes of the pre-synaptic neuron group
self.delay_step = int(delay/bm.get_dt())
self.pre_spike = bm.LengthDelay(pre.spike, self.delay_step)
# store the arrival time of the pre-synaptic spikes
self.spike_arrival_time = bm.Variable(bm.ones(self.pre.num) * -1e7)
# integral function
self.integral = bp.odeint(self.derivative, method=method)
def derivative(self, g, t, TT):
dg = self.alpha * TT * (1 - g) - self.beta * g
return dg
# for more details of how to run a simulation please see the tutorials in "Dynamics Simulation"
def show_syn_model(model):
pre = bp.neurons.LIF(1, V_rest=-60., V_reset=-60., V_th=-40.)
post = bp.neurons.LIF(1, V_rest=-60., V_reset=-60., V_th=-40.)
syn = model(pre, post, conn=bp.conn.One2One())
net = bp.Network(pre=pre, post=post, syn=syn)
runner = bp.DSRunner(net,
monitors=['pre.V', 'post.V', 'syn.g'],
inputs=['pre.input', 22.])
runner.run(100.)
fig, gs = bp.visualize.get_figure(1, 2, 3, 4)
fig.add_subplot(gs[0, 0])
bp.visualize.line_plot(runner.mon.ts, runner.mon['syn.g'], legend='syn.g')
fig.add_subplot(gs[0, 1])
bp.visualize.line_plot(runner.mon.ts, runner.mon['pre.V'], legend='pre.V')
bp.visualize.line_plot(runner.mon.ts, runner.mon['post.V'], legend='post.V', show=True)
Computation with Dense Connections#
Matrix-based synaptic computation is straightforward. Especially, when your models are connected densely, using matrix is highly efficient.
conn_mat#
Assume two neuron groups are connected through a fixed probability of 0.7.
conn = bp.conn.FixedProb(0.7)(pre_size=6, post_size=8)
Then you can create the connection matrix though conn.require("conn_mat"):
conn.require('conn_mat')
DeviceArray([[ True, False, True, True, True, True, False, True],
[ True, False, False, False, False, True, True, False],
[ True, True, True, True, False, True, True, True],
[ True, True, True, False, True, False, True, True],
[ True, False, True, True, True, True, False, False],
[ True, True, True, False, False, True, True, True]], dtype=bool)
conn_mat has the shape of (num_pre, num_post). Therefore, transforming the data with the pre-synaptic dimension into the date of the post-synaptic dimension is very easy. You just need make a matrix multiplication: brainpy.math.dot(pre_values, conn_mat) (\(\mathbb{R}^\mathrm{num\_pre} @ \mathbb{R}^\mathrm{(num\_pre, num\_post)} \to \mathbb{R}^\mathrm{num\_post}\)).
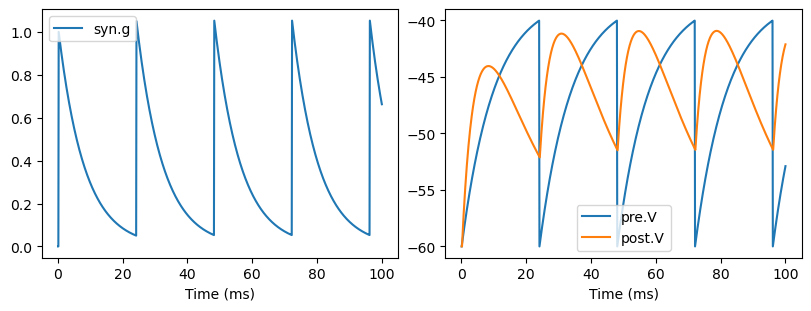
With the synaptic connection of conn_mat in above, we can define the exponential synapse model as the follows. It’s worthy to note that the evolution of states ouput onto the same post-synaptic neurons in exponential synapses can be superposed. This means we can declare the synapse variables with the shape of post-synaptic group, rather than the number of the total synapses.
class ExpConnMat(BaseExpSyn):
def __init__(self, *args, **kwargs):
super(ExpConnMat, self).__init__(*args, **kwargs)
# connection matrix
self.conn_mat = self.conn.require('conn_mat').astype(float)
# synapse gating variable
# -------
# NOTE: Here the synapse number is the same with
# the post-synaptic neuron number. This is
# different from the AMPA synapse.
self.g = bm.Variable(bm.zeros(self.post.num))
def update(self, tdi, x=None):
_t, _dt = tdi.t, tdi.dt
# pull the delayed pre spikes for computation
delayed_spike = self.pre_spike(self.delay_step)
# push the latest pre spikes into the bottom
self.pre_spike.update(self.pre.spike)
# integrate the synapse state
self.g.value = self.integral(self.g, _t, dt=_dt)
# update synapse states according to the pre spikes
post_sps = bm.dot(delayed_spike.astype(float), self.conn_mat)
self.g += post_sps
# get the post-synaptic current
self.post.input += self.g_max * self.g * (self.E - self.post.V)
show_syn_model(ExpConnMat)
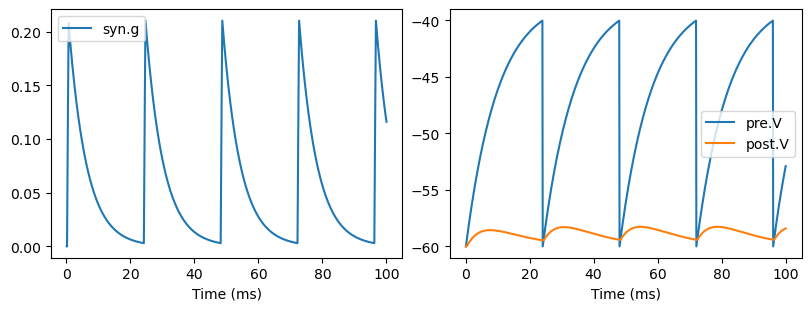
We can also use conn_mat to define an AMPA synapse model. Note here the shape of the synapse variable \(g\) is (num_pre, num_post), rather than self.post.num in the above exponential synapse model. This is because the synaptic states of AMPA model can not be superposed.
class AMPAConnMat(BaseAMPASyn):
def __init__(self, *args, **kwargs):
super(AMPAConnMat, self).__init__(*args, **kwargs)
# connection matrix
self.conn_mat = self.conn.require('conn_mat').astype(float)
# synapse gating variable
# -------
# NOTE: Here the synapse shape is (num_pre, num_post),
# in contrast to the ExpConnMat
self.g = bm.Variable(bm.zeros((self.pre.num, self.post.num)))
def update(self, tdi, x=None):
_t, _dt = tdi.t, tdi.dt
# pull the delayed pre spikes for computation
delayed_spike = self.pre_spike(self.delay_step)
# push the latest pre spikes into the bottom
self.pre_spike.update(self.pre.spike)
# get the time of pre spikes arrive at the post synapse
self.spike_arrival_time.value = bm.where(delayed_spike, _t, self.spike_arrival_time)
# get the neurotransmitter concentration at the current time
TT = ((_t - self.spike_arrival_time) < self.T_duration) * self.T
# integrate the synapse state
TT = TT.reshape((-1, 1)) * self.conn_mat # NOTE: only keep the concentrations
# on the invalid connections
self.g.value = self.integral(self.g, _t, TT, dt=_dt)
# get the post-synaptic current
g_post = self.g.sum(axis=0)
self.post.input += self.g_max * g_post * (self.E - self.post.V)
show_syn_model(AMPAConnMat)
Special connections#
Sometimes, we can define some synapse models with special connection types, such as all-to-all connection, or one-to-one connection. For these special situations, even the connection information can be ignored, i.e., we do not need conn_mat or other structures anymore.
Assume the pre-synaptic group connects to the post-synaptic group with a all-to-all fashion. Then, exponential synapse model can be defined as,
class ExpAll2All(BaseExpSyn):
def __init__(self, *args, **kwargs):
super(ExpAll2All, self).__init__(*args, **kwargs)
# synapse gating variable
# -------
# The synapse variable has the shape of the post-synaptic group
self.g = bm.Variable(bm.zeros(self.post.num))
def update(self, tdi, x=None):
_t, _dt = tdi.t, tdi.dt
delayed_spike = self.pre_spike(self.delay_step)
self.pre_spike.update(self.pre.spike)
self.g.value = self.integral(self.g, _t, dt=_dt)
self.g += delayed_spike.sum() # NOTE: HERE is the difference
self.post.input += self.g_max * self.g * (self.E - self.post.V)
show_syn_model(ExpAll2All)

Similarly, the AMPA synapse model can be defined as
class AMPAAll2All(BaseAMPASyn):
def __init__(self, *args, **kwargs):
super(AMPAAll2All, self).__init__(*args, **kwargs)
# synapse gating variable
# -------
# The synapse variable has the shape of the post-synaptic group
self.g = bm.Variable(bm.zeros((self.pre.num, self.post.num)))
def update(self, tdi, x=None):
_t, _dt = tdi.t, tdi.dt
delayed_spike = self.pre_spike(self.delay_step)
self.pre_spike.update(self.pre.spike)
self.spike_arrival_time.value = bm.where(delayed_spike, _t, self.spike_arrival_time)
TT = ((_t - self.spike_arrival_time) < self.T_duration) * self.T
TT = TT.reshape((-1, 1)) # NOTE: here is the difference
self.g.value = self.integral(self.g, _t, TT, dt=_dt)
g_post = self.g.sum(axis=0) # NOTE: here is also different
self.post.input += self.g_max * g_post * (self.E - self.post.V)
show_syn_model(AMPAAll2All)

Actually, the synaptic computation with these special connections can be very efficient! A concrete example please see a decision making spiking model in BrainPy-Examples. This implementation achieve at least four times acceleration comparing to the implementation in other frameworks.
Computation with Sparse Connections#
However, in the real neural system, the neurons are connected sparsely in essence.
Imaging you want to connect 10,000 pre-synaptic neurons to 10,000 post-synaptic neurons with a 10% random connection probability. Using matrix, you need \(10^8\) floats to save the synaptic state, and at each update step, you need do computation on \(10^8\) floats. Actually, the number of synapses you really connect is only \(10^7\). See, there is a huge memory waste and computing resource inefficiency. Moreover, at the given time \(\mathrm{\_t}\), the number of pre-synaptic neurons in the spiking state is small on average. This means we have made many useless computations when defining synaptic computations with matrix-based connections (zeros dot connection matrix results in zeros).
Therefore, we need new ways to define synapse models. Specifically, we use vectors to store the connected neuron indices, like the pre_ids and post_ids (see Synaptic Connections).
In the below, we assume you have learned the synaptic connection types detailed in the tutorial of Synaptic Connections.
The pre2post operator#
A notable difference of brain dynamics models from the deep learning is that they are sparse and event-driven. In order to support this significant different kind of computations, BrainPy has built many useful operators. In this section, we talk about a set of operators needed in pre2post computations.
Note before we have said that exponential synapse model can make computations at the dimension of the post-synaptic group. Therefore, we can directly transform the pre-synaptic data into the data of the post-synaptic shape. brainpy.math.pre2post_event_sum(events, pre2post, post_num, values) can satisfy your requirements. This operator needs the synaptic structure of pre2post (a tuple contains the post_ids and idnptr of pre-synaptic neurons).
If values is a scalar, pre2post_event_sum is equivalent to:
post_val = np.zeros(post_num)
post_ids, idnptr = pre2post
for i in range(pre_num):
if events[i]:
for j in range(idnptr[i], idnptr[i+1]):
post_val[post_ids[i]] += values
If values is a vector, pre2post_event_sum is equivalent to:
post_val = np.zeros(post_num)
post_ids, idnptr = pre2post
for i in range(pre_num):
if events[i]:
for j in range(idnptr[i], idnptr[i+1]):
post_val[post_ids[i]] += values[j]
With this operator, exponential synapse model can be defined as:
class ExpSparse(BaseExpSyn):
def __init__(self, *args, **kwargs):
super(ExpSparse, self).__init__(*args, **kwargs)
# connections
self.pre2post = self.conn.require('pre2post')
# synapse variable
self.g = bm.Variable(bm.zeros(self.post.num))
def update(self, tdi, x=None):
_t, _dt = tdi.t, tdi.dt
delayed_spike = self.pre_spike(self.delay_step)
self.pre_spike.update(self.pre.spike)
self.g.value = self.integral(self.g, _t, dt=_dt)
# NOTE: update synapse states according to the pre spikes
post_sps = bm.pre2post_event_sum(delayed_spike, self.pre2post, self.post.num, 1.)
self.g += post_sps
self.post.input += self.g_max * self.g * (self.E - self.post.V)
show_syn_model(ExpSparse)

This model will be very efficient when your synapses are connected sparsely.
The pre2syn and syn2post operators#
However, for AMPA synapse model, the pre-synaptic values can not be directly transformed into the post-synaptic dimensional data. Therefore, we need to first change the pre data into the data of the synapse dimension, then transform the synapse-dimensional data into the post-dimensional data.
Therefore, the core problem of synaptic computation is how to convert values among different shape of arrays. Specifically, in the above AMPA synapse model, we have three kinds of array shapes (see the following figure): arrays with the dimension of pre-synaptic group, arrays of the dimension of post-synaptic group, and arrays with the shape of synaptic connections. Converting the pre-synaptic spiking state into the synaptic state and grouping the synaptic variable as the post-synaptic current value are central problems of synaptic computation.
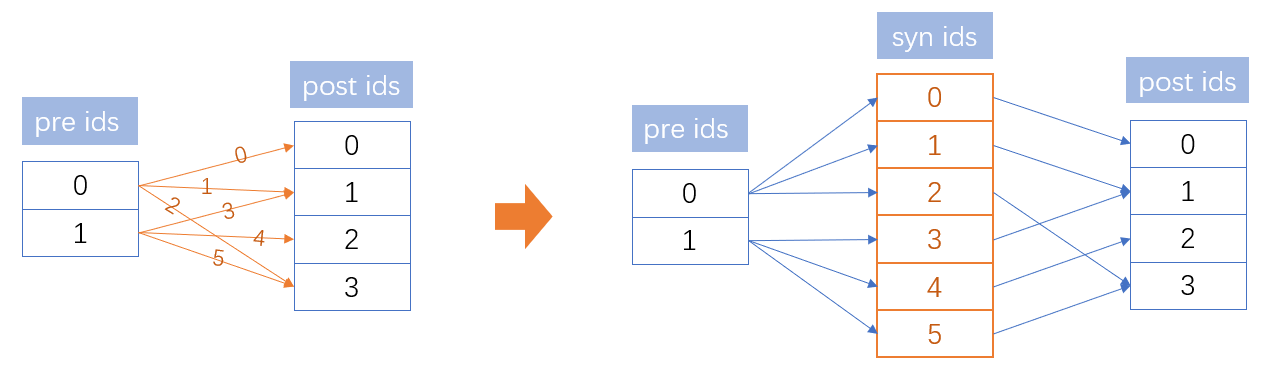
Here BrainPy provides two operators brainpy.math.pre2syn(pre_values, pre_ids) and brainpy.math.syn2post(syn_values, post_ids, post_num) to convert vectors among different dimensions.
brainpy.math.pre2syn()receives two arguments: “pre_values” (the variable of the pre-synaptic dimension) and “pre_ids” (the connected pre-synaptic neuron index).brainpy.math.syn2post()receives three arguments: “syn_values” (the variable with the synaptic size), “post_ids” (the connected post-synaptic neuron index) and “post_num” (the number of the post-synaptic neurons).
Based on these two operators, we can define the AMPA synapse model as:
class AMPASparse(BaseAMPASyn):
def __init__(self, *args, **kwargs):
super(AMPASparse, self).__init__(*args, **kwargs)
# connection matrix
self.pre_ids, self.post_ids = self.conn.require('pre_ids', 'post_ids')
# synapse gating variable
# -------
# NOTE: Here the synapse shape is (num_syn,)
self.g = bm.Variable(bm.zeros(len(self.pre_ids)))
def update(self, tdi, x=None):
_t, _dt = tdi.t, tdi.dt
delayed_spike = self.pre_spike(self.delay_step)
self.pre_spike.update(self.pre.spike)
# get the time of pre spikes arrive at the post synapse
self.spike_arrival_time.value = bm.where(delayed_spike, _t, self.spike_arrival_time)
# get the arrival time with the synapse dimension
arrival_times = bm.pre2syn(self.spike_arrival_time, self.pre_ids)
# get the neurotransmitter concentration at the current time
TT = ((_t - arrival_times) < self.T_duration) * self.T
# integrate the synapse state
self.g.value = self.integral(self.g, _t, TT, dt=_dt)
# get the post-synaptic current
g_post = bm.syn2post(self.g, self.post_ids, self.post.num)
self.post.input += self.g_max * g_post * (self.E - self.post.V)
show_syn_model(AMPASparse)

We hope this tutorial will help users build synapse models in a more efficient way.

