Simulation with DSRunner#
@Tianqiu Zhang @Chaoming Wang @Xiaoyu Chen
The convenient simulation interface for dynamical systems in BrainPy is implemented by brainpy.dyn.DSRunner. It can simulate various levels of models including channels, neurons, synapses and systems. In this tutorial, we will introduce how to use brainpy.dyn.DSRunner in detail.
import brainpy as bp
import brainpy.math as bm
bm.set_platform('cpu')
[Taichi] version 1.7.0, llvm 15.0.1, commit 37b8e80c, win, python 3.11.5
bp.__version__
'2.4.6'
Initializing a DSRunner#
Generally, we can initialize a runner for dynamical systems with the format of:
runner = DSRunner(
target=instance_of_dynamical_system,
inputs=inputs_for_target_DynamicalSystem,
monitors=interested_variables_to_monitor,
jit=enable_jit_or_not,
progress_bar=report_the_running_progress,
numpy_mon_after_run=transform_into_numpy_ndarray
)
In which
targetspecifies the model to be simulated. It must an instance of brainpy.DynamicalSystem.inputsis used to define the input operations for specific variables.It should be the format of
[(target, value, [type, operation])], wheretargetis the input target,valueis the input value,typeis the input type (such as “fix”, “iter”, “func”),operationis the operation for inputs (such as “+”, “-”, “*”, “/”, “=”). Also, if you want to specify multiple inputs, just give multiple(target, value, [type, operation]), such as[(target1, value1), (target2, value2)].It can also be a function, which is used to manually specify the inputs for the target variables. This input function should receive one argument
tdiwhich contains the shared arguments like timet, time stepdt, and indexi.
monitorsis used to define target variables in the model. During the simulation, the history values of the monitored variables will be recorded. It can also to monitor variables by callable functions and it should be adict. Thekeyshould be a string for later retrieval byrunner.mon[key]. Thevalueshould be a callable function which receives an argument:tdt.jitdetermines whether to use JIT compilation during the simulation.progress_bardetermines whether to use progress bar to report the running progress or not.numpy_mon_after_rundetermines whether to transform the JAX arrays into numpy ndarray or not when the network finishes running.
Running a DSRunner#
After initialization of the runner, users can call .run() function to run the simulation. The format of function .run() is showed as follows:
runner.run(duration=simulation_time_length,
inputs=input_data,
reset_state=whether_reset_the_model_states,
shared_args=shared_arguments_across_different_layers,
progress_bar=report_the_running_progress,
eval_time=evaluate_the_running_time
)
In which
durationis the simulation time length.inputsis the input data. Ifinputs_are_batching=True,inputsmust be a PyTree of data with two dimensions:(num_sample, num_time, ...). Otherwise, theinputsshould be a PyTree of data with one dimension:(num_time, ...).reset_statedetermines whether to reset the model states.shared_argsis shared arguments across different layers. All the layers can access the elements inshared_args.progress_bardetermines whether to use progress bar to report the running progress or not.eval_timedetermines whether to evaluate the running time.
Here we define an E/I balance network as the simulation model.
class EINet(bp.Network):
def __init__(self, scale=1.0, method='exp_auto'):
super(EINet, self).__init__()
# network size
num_exc = int(3200 * scale)
num_inh = int(800 * scale)
# neurons
pars = dict(V_rest=-60., V_th=-50., V_reset=-60., tau=20., tau_ref=5.)
self.E = bp.neurons.LIF(num_exc, **pars, method=method)
self.I = bp.neurons.LIF(num_inh, **pars, method=method)
# synapses
prob = 0.1
we = 0.6 / scale / (prob / 0.02) ** 2 # excitatory synaptic weight (voltage)
wi = 6.7 / scale / (prob / 0.02) ** 2 # inhibitory synaptic weight
self.E2E = bp.synapses.Exponential(self.E, self.E, bp.conn.FixedProb(prob),
output=bp.synouts.COBA(E=0.), g_max=we,
tau=5., method=method)
self.E2I = bp.synapses.Exponential(self.E, self.I, bp.conn.FixedProb(prob),
output=bp.synouts.COBA(E=0.), g_max=we,
tau=5., method=method)
self.I2E = bp.synapses.Exponential(self.I, self.E, bp.conn.FixedProb(prob),
output=bp.synouts.COBA(E=-80.), g_max=wi,
tau=10., method=method)
self.I2I = bp.synapses.Exponential(self.I, self.I, bp.conn.FixedProb(prob),
output=bp.synouts.COBA(E=-80.), g_max=wi,
tau=10., method=method)
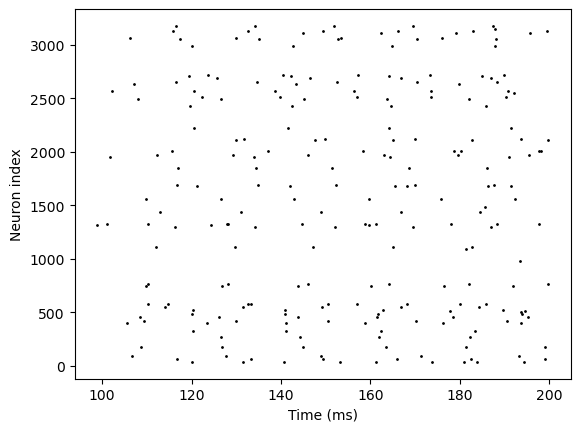
Then we will wrap it into DSRunner for dynamic simulation. brainpy.DSRunner aims to provide model simulation with an outstanding performance. It takes advantage of the structural loop primitive to lower the model onto the XLA devices.
# instantiate EINet
net = EINet()
# initialize DSRunner
runner = bp.DSRunner(target=net,
monitors=['E.spike'],
inputs=[('E.input', 20.), ('I.input', 20.)],
jit=True)
# run the simulation
runner.run(duration=1000.)
bp.visualize.raster_plot(runner.mon.ts, runner.mon['E.spike'])
We have run a simple example of using DSRunner, but there are many advanced usages despite this. Next we will formally introduce two main aspects that will be used frequently in DSRunner: monitors and inputs.
Monitors in DSRunner#
In BrainPy, any instance of brainpy.DSRunner has a built-in monitor. Users can set up a monitor when initializing a runner. There are multiple methods to initialize a monitor. The first method is to initialize a monitor is through a list of strings.
Initialization with a list of strings#
# initialize monitor through a list of strings
runner1 = bp.DSRunner(target=net,
monitors=['E.spike', 'E.V', 'I.spike', 'I.V'], # 4 elements in monitors
inputs=[('E.input', 20.), ('I.input', 20.)],
jit=True)
where all the strings corresponds to the name of the variables in the EI network:
net.E.V, net.E.spike
(Variable(value=Array([-55.740116, -54.815796, -56.08671 , ..., -53.89189 , -53.306213,
-53.387295]),
dtype=float32),
Variable(value=Array([False, False, False, ..., False, False, False]), dtype=bool))
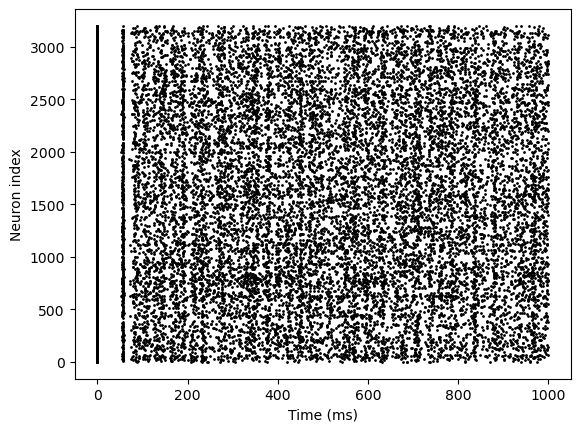
Once we call the runner with a given time duration, the monitor will automatically record the variable evolutions in the corresponding models. Afterwards, users can access these variable trajectories by using .mon.[variable_name]. The default history times .mon.ts will also be generated after the model finishes its running. Let’s see an example.
runner1.run(100.)
bp.visualize.raster_plot(runner1.mon.ts, runner1.mon['E.spike'], show=True)
Initialization with index specification#
The second method is similar to the first one, with the difference that the index specification is added. Index specification means users only monitor the specific neurons and ignore all the other neurons. Sometimes we do not care about all the contents in a variable. We may be only interested in the values at the certain indices. Moreover, for a huge network with a long-time simulation, monitors will consume a large part of RAM. Therefore, monitoring variables only at the selected indices will be more applicable. BrainPy supports monitoring a part of elements in a Variable with the format of tuple like this:
# initialize monitor through a list of strings with index specification
runner2 = bp.DSRunner(target=net,
monitors=[('E.spike', [1, 2, 3]), # monitor values of Variable at index of [1, 2, 3]
'E.V'], # monitor all values of Variable 'V'
inputs=[('E.input', 20.), ('I.input', 20.)],
jit=True)
runner2.run(100.)
print('The monitor shape of "E.V" is (run length, variable size) = {}'.format(runner2.mon['E.V'].shape))
print('The monitor shape of "E.spike" is (run length, index size) = {}'.format(runner2.mon['E.spike'].shape))
The monitor shape of "E.V" is (run length, variable size) = (1000, 3200)
The monitor shape of "E.spike" is (run length, index size) = (1000, 3)
Explicit monitor target#
The third method is to use a dict with the explicit monitor target. Users can access model instance and get certain variables as monitor target:
# initialize monitor through a dict with the explicit monitor target
runner3 = bp.DSRunner(target=net,
monitors={'spike': net.E.spike, 'V': net.E.V},
inputs=[('E.input', 20.), ('I.input', 20.)],
jit=True)
runner3.run(100.)
print('The monitor shape of "V" is = {}'.format(runner3.mon['V'].shape))
print('The monitor shape of "spike" is = {}'.format(runner3.mon['spike'].shape))
The monitor shape of "V" is = (1000, 3200)
The monitor shape of "spike" is = (1000, 3200)
Explicit monitor target with index specification#
The fourth method is similar to the third one, with the difference that the index specification is added:
# initialize monitor through a dict with the explicit monitor target
runner4 = bp.DSRunner(target=net,
monitors={'E.spike': (net.E.spike, [1, 2]), # monitor values of Variable at index of [1, 2]
'E.V': net.E.V}, # monitor all values of Variable 'V'
inputs=[('E.input', 20.), ('I.input', 20.)],
jit=True)
runner4.run(100.)
print('The monitor shape of "E.V" is = {}'.format(runner4.mon['E.V'].shape))
print('The monitor shape of "E.spike" is = {}'.format(runner4.mon['E.spike'].shape))
The monitor shape of "E.V" is = (1000, 3200)
The monitor shape of "E.spike" is = (1000, 2)
In spite of the four methods mentioned above, BrainPy also provides users a convenient parameter to monitor more complicate variables: fun_monitor. Users can use a function to describe monitor and pass to fun_monitor. fun_monitor must be a dict and the key should be a string for the later retrieval by runner.mon[key], the value should be a callable function which receives an arguments: tdi. The format of fun_monitor is shown as below:
fun_monitor = {'key_name': lambda tdi: body_func(tdi)}
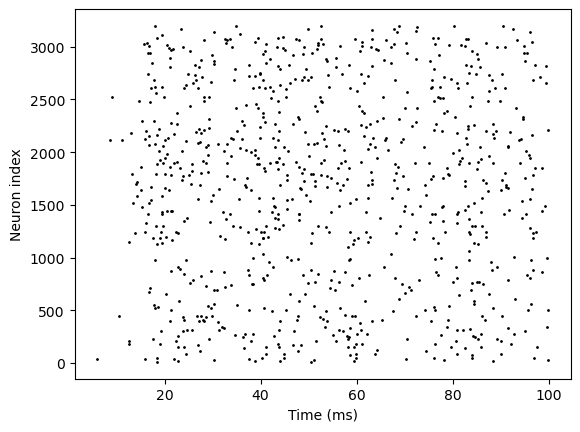
Here we monitor a variable that
runner5 = bp.DSRunner(target=net,
monitors={'E-I.spike': lambda: bm.concatenate((net.E.spike, net.I.spike), axis=0)},
inputs=[('E.input', 20.), ('I.input', 20.)],
jit=True)
runner5.run(100.)
bp.visualize.raster_plot(runner5.mon.ts, runner5.mon['E-I.spike'])
D:\codes\projects\brainpy-chaoming0625\brainpy\_src\runners.py:36: UserWarning:
From brainpy>=2.4.3, input() and monitor() function no longer needs to receive a global shared argument.
Instead of using:
def f_input_or_monitor(tdi):
...
Please use:
def f_input_or_monitor():
t = bp.share['t']
...
warnings.warn(_input_deprecate_msg, UserWarning)
Inputs in DSRunner#
In brain dynamics simulation, various inputs are usually given to different units of the dynamical system. In BrainPy, inputs can be specified to runners for dynamical systems. The aim of inputs is to mimic the input operations in experiments like Transcranial Magnetic Stimulation (TMS) and patch clamp recording.
inputs should have the format like (target, value, [type, operation]), where
targetis the target variable to inject the input.valueis the input value. It can be a scalar, a tensor, or a iterable object/function.typeis the type of the input value. It support two types of input:fixanditer. The first one means that the data is static; the second one denotes the data can be iterable, no matter whether the input value is a tensor or a function. Theitertype must be explicitly stated.operationis the input operation on the target variable. It should be set as one of{ + , - , * , / , = }, and if users do not provide this item explicitly, it will be set to ‘+’ by default, which means that the target variable will be updated asval = val + input.
Users can also give multiple inputs for different target variables, like:
inputs=[(target1, value1, [type1, op1]),
(target2, value2, [type2, op2]),
... ]
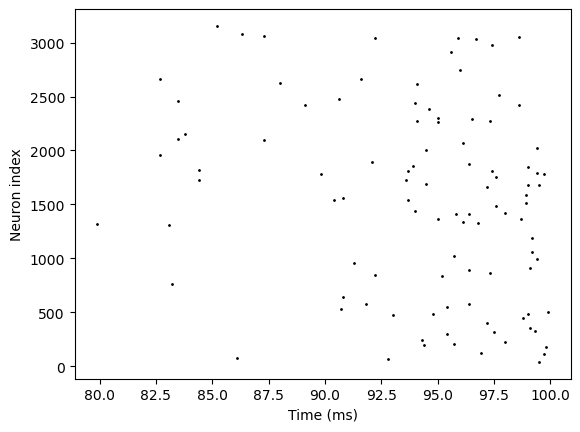
Static inputs#
The first example is providing static inputs. The excitation and inhibition neurons all receive the same current intensity:
runner6 = bp.DSRunner(target=net,
monitors=['E.spike'],
inputs=[('E.input', 20.), ('I.input', 20.)], # static inputs
jit=True)
runner6.run(100.)
bp.visualize.raster_plot(runner6.mon.ts, runner6.mon['E.spike'])
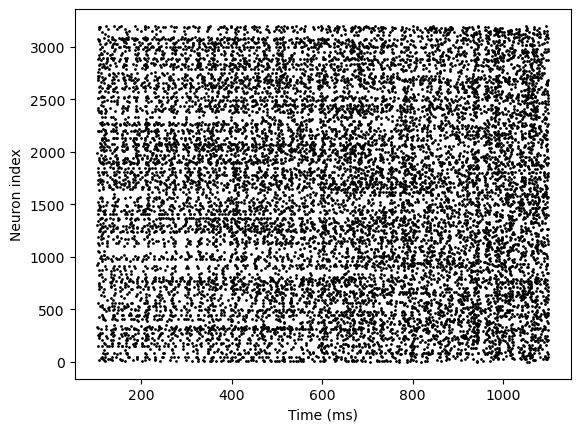
Iterable inputs#
The second example is providing iterable inputs. Users need to set type=iter and pass an iterable object or function into value:
I, length = bp.inputs.section_input(values=[0, 20., 0],
durations=[100, 1000, 100],
return_length=True,
dt=0.1)
runner7 = bp.DSRunner(target=net,
monitors=['E.spike'],
inputs=[('E.input', I, 'iter'), ('I.input', I, 'iter')], # iterable inputs
jit=True)
runner7.run(length)
bp.visualize.raster_plot(runner7.mon.ts, runner7.mon['E.spike'])
By examples given above, users can easily understand the usage of inputs parameters. Similar to monitors, inputs can also be more complicate as a function form. BrainPy provides fun_inputs to receive the customized functional inputs created by users.
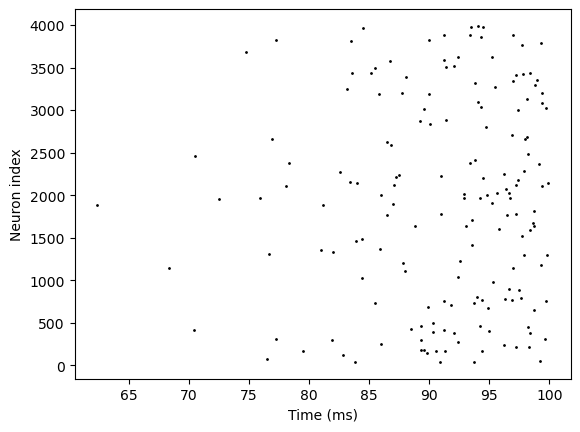
def set_input():
net.E.input[:] = 20
net.I.input[:] = 20.
runner8 = bp.DSRunner(target=net,
monitors=['E.spike'],
inputs=set_input, # functional inputs
jit=True)
runner8.run(200.)
bp.visualize.raster_plot(runner8.mon.ts, runner8.mon['E.spike'])
D:\codes\projects\brainpy-chaoming0625\brainpy\_src\runners.py:36: UserWarning:
From brainpy>=2.4.3, input() and monitor() function no longer needs to receive a global shared argument.
Instead of using:
def f_input_or_monitor(tdi):
...
Please use:
def f_input_or_monitor():
t = bp.share['t']
...
warnings.warn(_input_deprecate_msg, UserWarning)