How to customize a synapse#
import numpy as np
import brainpy as bp
import brainpy.math as bm
import matplotlib.pyplot as plt
Preparations#
brainpy.dyn.ProjAlignPostMg2#
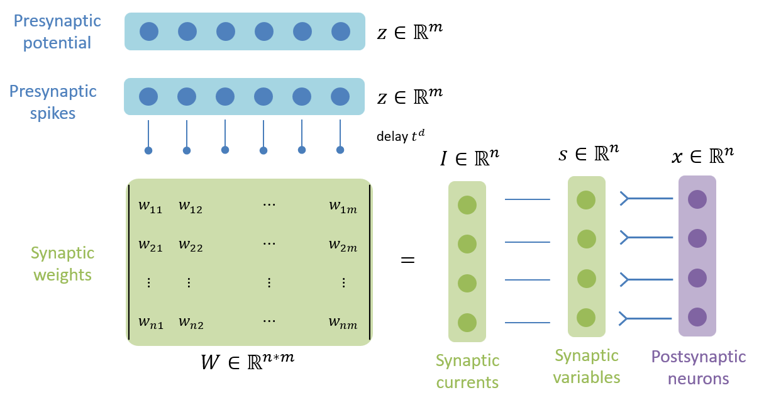
Synaptic projection which defines the synaptic computation with the dimension of postsynaptic neuron group.
brainpy.dyn.ProjAlignPostMg2(
pre,
delay,
comm,
syn,
out,
post
)
pre (JointType[DynamicalSystem, AutoDelaySupp]): The pre-synaptic neuron group.delay (Union[None, int, float]): The synaptic delay.comm (DynamicalSystem): The synaptic communication.syn (ParamDescInit): The synaptic dynamics.out (ParamDescInit): The synaptic output.post (DynamicalSystem)The post-synaptic neuron group.
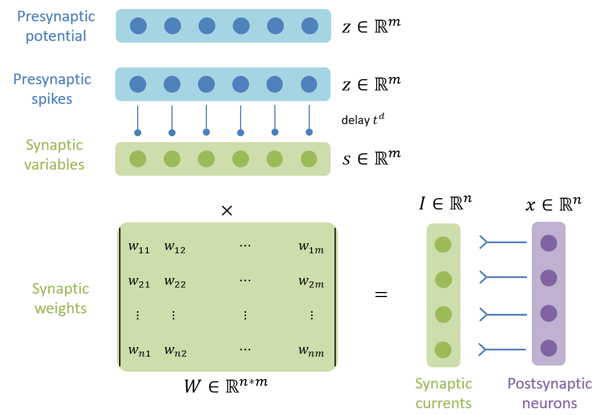
brainpy.dyn.ProjAlignPreMg2#
Synaptic projection which defines the synaptic computation with the dimension of presynaptic neuron group.
brainpy.dyn.ProjAlignPreMg2(
pre,
delay,
syn,
comm,
out,
post
)
pre (JointType[DynamicalSystem, AutoDelaySupp]): The pre-synaptic neuron group.delay (Union[None, int, float]): The synaptic delay.syn (ParamDescInit): The synaptic dynamics.comm (DynamicalSystem): The synaptic communication.out (ParamDescInit): The synaptic output.post (DynamicalSystem)The post-synaptic neuron group.
Synapse dynamics#
Exponential Model#
The single exponential decay synapse model assumes the release of neurotransmitter, its diffusion across the cleft, the receptor binding, and channel opening all happen very quickly, so that the channels instantaneously jump from the closed to the open state. Therefore, its expression is given by
where \(\tau\) is the time constant, \(t_0\) is the time of the pre-synaptic spike, \(\bar{g}_{\mathrm{syn}}\) is the maximal conductance.
The corresponding differential equation:
Only Exponential synapse model can use the AlignPost.
class Exponen(bp.dyn.SynDyn, bp.mixin.AlignPost):
def __init__(self, size, tau):
super().__init__(size)
# parameters
self.tau = tau
# variables
self.g = bm.Variable(bm.zeros(self.num))
# integral
self.integral = bp.odeint(lambda g, t: -g/tau, method='exp_auto')
def update(self, pre_spike=None):
self.g.value = self.integral(g=self.g.value, t=bp.share['t'], dt=bp.share['dt'])
if pre_spike is not None:
self.add_current(pre_spike)
return self.g.value
def add_current(self, x): # specical for bp.mixin.AlignPost
self.g += x
def return_info(self):
return self.g
AMPA model#
AMAP synapse is described by a differential equation.
Where \(\alpha [T]\) denotes the transition probability from state \((1-s)\) to state \((s)\); and \(\beta\) represents the transition probability of the other direction. \(\alpha=0.98\) is the binding constant. \(\beta=.18\) is the unbinding constant. \(T=.5\, mM\) is the neurotransmitter concentration, and has the duration of 0.5 ms.
class AMPA(bp.dyn.SynDyn):
def __init__(self, size, alpha= 0.98, beta=0.18, T=0.5, T_dur=0.5):
super().__init__(size=size)
# parameters
self.alpha = alpha
self.beta = beta
self.T = T
self.T_duration = T_dur
# functions
self.integral = bp.odeint(method='exp_auto', f=self.dg)
# variables
self.g = bm.Variable(bm.zeros(self.num))
self.spike_arrival_time = bm.Variable(bm.ones(self.num) * -1e7)
def dg(self, g, t, TT):
return self.alpha * TT * (1 - g) - self.beta * g
def update(self, pre_spike):
self.spike_arrival_time.value = bm.where(pre_spike, bp.share['t'], self.spike_arrival_time)
TT = ((bp.share['t'] - self.spike_arrival_time) < self.T_duration) * self.T
self.g.value = self.integral(self.g, bp.share['t'], TT, bp.share['dt'])
return self.g.value
def return_info(self):
return self.g
Synapse outputs#
COBA#
Given the synaptic conductance, the COBA model outputs the post-synaptic current with
class COBA(bp.dyn.SynOut):
def __init__(self, E):
super().__init__()
self.E = E
def update(self, conductance, potential):
return conductance * (self.E - potential)
CUBA#
Given the conductance, this model outputs the post-synaptic current with a identity function:
class CUBA(bp.dyn.SynOut):
def __init__(self, E):
super().__init__()
self.E = E
def update(self, conductance, potential):
return conductance
Mg Blocking#
The voltage dependence is due to the blocking of the pore of the NMDA receptor from the outside by a positively charged magnesium ion. The channel is nearly completely blocked at resting potential, but the magnesium block is relieved if the cell is depolarized. The fraction of channels \(B(V)\) that are not blocked by magnesium can be fitted to
Here, \([{Mg}^{2+}]_{o}\) is the extracellular magnesium concentration, usually 1 mM.
If we make the approximation that the magnesium block changes instantaneously with voltage and is independent of the gating of the channel, the net NMDA receptor-mediated synaptic current is given by
where \(V(t)\) is the post-synaptic neuron potential, \(E\) is the reversal potential.
class MgBlock(bp.dyn.SynOut):
def __init__(self, E= 0., cc_Mg= 1.2, alpha= 0.062, beta= 3.57):
super().__init__()
self.E = E
self.cc_Mg = cc_Mg
self.alpha = alpha
self.beta = beta
def update(self, conductance, potential):
return conductance * (self.E - potential) / (1 + self.cc_Mg / self.beta * bm.exp(-self.alpha * potential))
Synaptic communications#
Masked matrix#
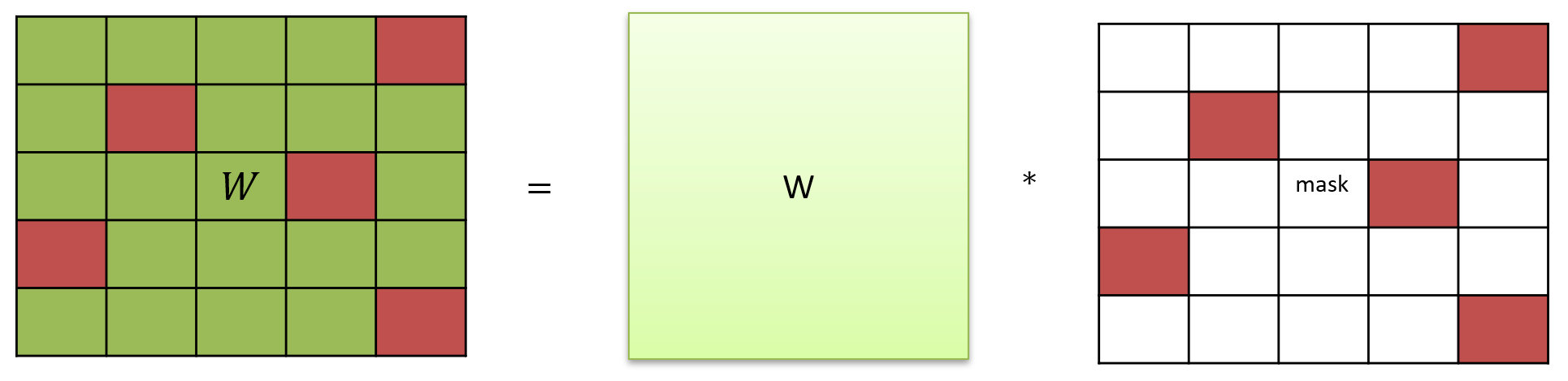
class MaskedLinear(bp.dnn.Layer):
def __init__(self, conn, weight):
super().__init__()
# connection and weight
weight = bp.init.parameter(weight, (conn.pre_num, conn.post_num))
if isinstance(self.mode, bm.TrainingMode):
weight = bm.TrainVar(weight)
self.weight = weight
# connection
self.conn = conn
self.mask = bm.sharding.partition(self.conn.require('conn_mat'))
def update(self, x):
return x @ (self.weight * self.mask)
Examples#
class SimpleNet(bp.DynSysGroup):
def __init__(self, proj, g_max=1.):
super().__init__()
self.pre = bp.dyn.SpikeTimeGroup(1, indices=(0, 0, 0, 0), times=(10., 30., 50., 70.))
self.post = bp.dyn.LifRef(1, V_rest=-60., V_th=-50., V_reset=-60., tau=20., tau_ref=5.,
V_initializer=bp.init.Constant(-60.))
self.syn = proj(self.pre, self.post, delay=None, prob=1., g_max=g_max)
def update(self):
self.pre()
self.syn()
self.post()
# monitor the following variables
conductance = self.syn.proj.refs['syn'].g
current = self.post.sum_inputs(self.post.V)
return conductance, current, self.post.V
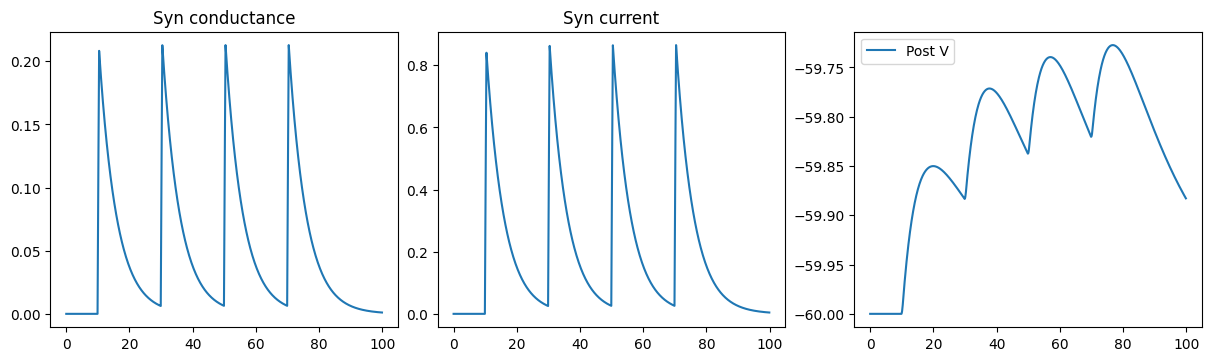
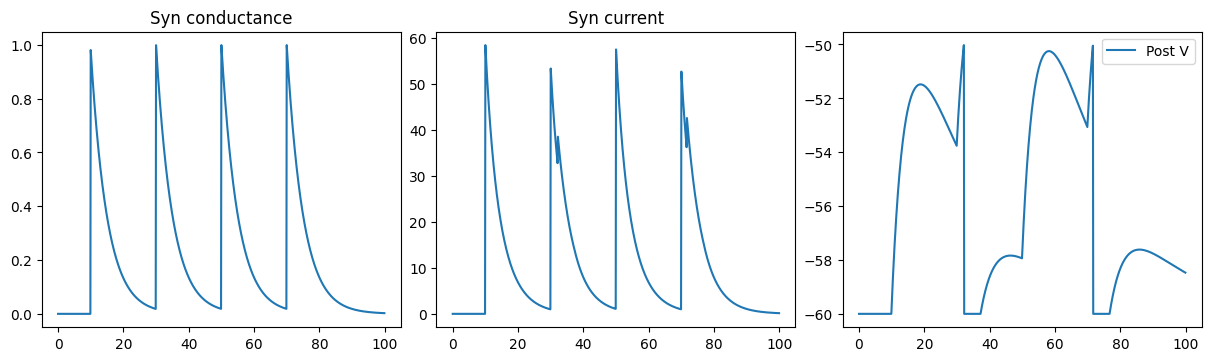
def run_a_net(net):
indices = np.arange(1000) # 100 ms
conductances, currents, potentials = bm.for_loop(net.step_run, indices, progress_bar=True)
# --- similar to:
# runner = bp.DSRunner(net)
# conductances, currents, potentials = runner.run(100.)
ts = indices * bm.get_dt()
fig, gs = bp.visualize.get_figure(1, 3, 3.5, 4)
fig.add_subplot(gs[0, 0])
plt.plot(ts, conductances)
plt.title('Syn conductance')
fig.add_subplot(gs[0, 1])
plt.plot(ts, currents)
plt.title('Syn current')
fig.add_subplot(gs[0, 2])
plt.plot(ts, potentials, label='Post V')
plt.legend()
plt.show()
Exponential COBA#
class ExponCOBA(bp.Projection):
def __init__(self, pre, post, delay, prob, g_max, tau=5., E=0.):
super().__init__()
self.proj = bp.dyn.ProjAlignPostMg2(
pre=pre,
delay=delay,
comm=MaskedLinear(bp.conn.FixedProb(prob, pre=pre.num, post=post.num), g_max),
syn=Exponen.desc(post.num, tau=tau),
out=COBA.desc(E=E),
post=post,
)
run_a_net(SimpleNet(ExponCOBA))
AMPA NMDA#
class AMPA_NMDA(bp.Projection):
def __init__(self, pre, post, delay, prob, g_max, E=0.):
super().__init__()
self.proj = bp.dyn.ProjAlignPreMg2(
pre=pre,
delay=delay,
syn=AMPA.desc(post.num),
comm=MaskedLinear(bp.conn.FixedProb(prob, pre=pre.num, post=post.num), g_max),
out=MgBlock(E=E),
post=post,
)
run_a_net(SimpleNet(AMPA_NMDA))