Surrogate gradient#
In recent years, spiking neural networks (SNNs) show their promising advantages in energy efficiency, fault tolerance, and biological plausibility. However, SNNs are difficult to train using standard gradient descent methods because their activation functions are discontinuous and have zero gradients almost everywhere. The commonly used way is to replace the non-differentiable spiking function with the surrogate gradient function. A surrogate gradient function is a smooth function that approximates the derivative of the activation function and allows gradient-based learning algorithms to be applied to SNNs.
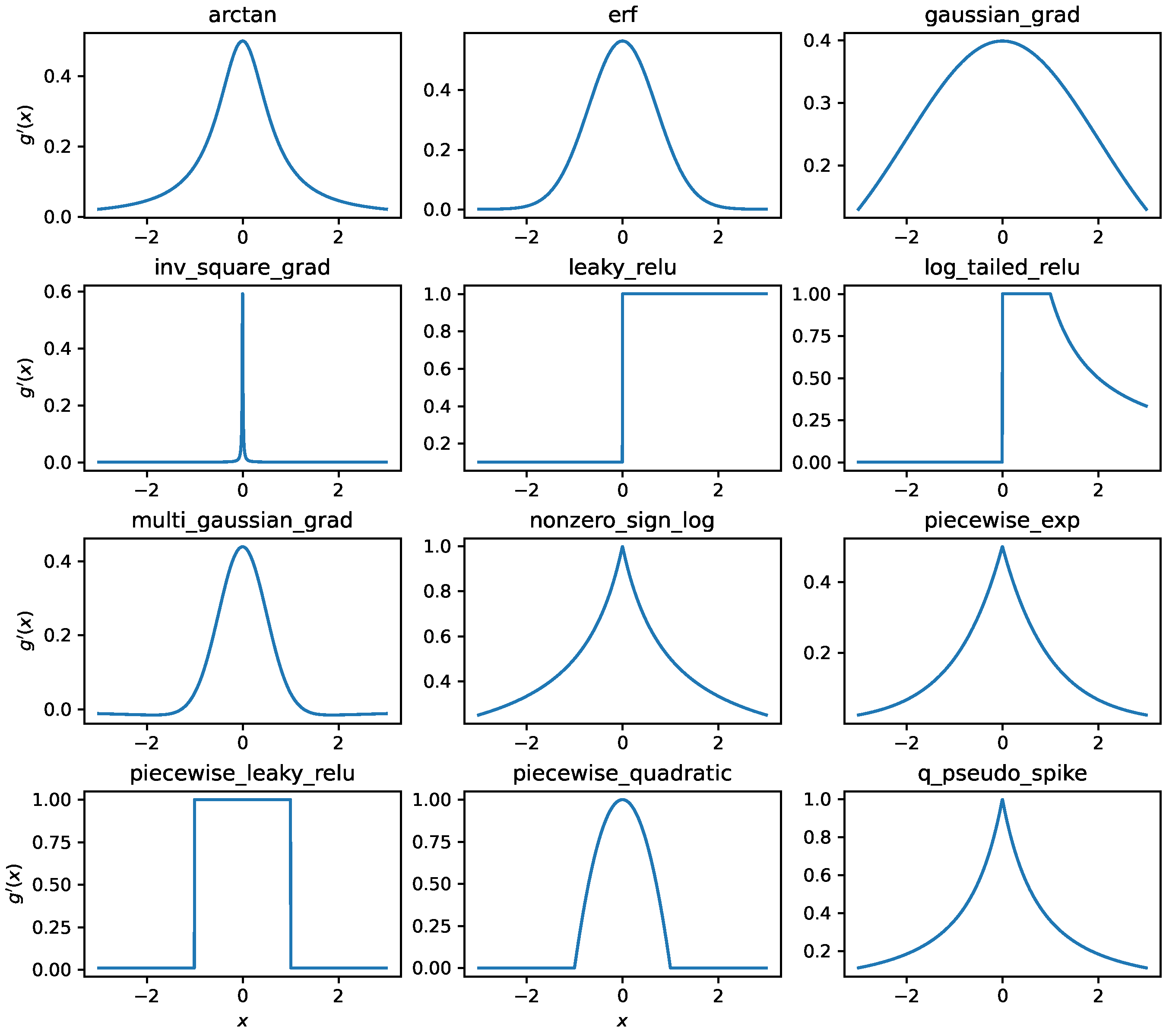
BrainPy provides multiple surrogate gradient functions with different properties of smoothness, boundedness, and biological plausibility. The full list is shown in Table below, and for the example of the surrogate gradient function please see the Figure Below.
In practice, users can use these surrogate gradient functions as parameters in neuron models. For example, in the leaky integrate-and-fire (LIF) neuron model brainpy.neurons.LIF, use can use:
model = brainpy.neurons.LIF (... , spike_fun=<surrogate function >)
Name |
Implementation |
|---|---|
Sigmoid function |
|
Piecewise quadratic function |
|
Piecewise exponential function |
|
Soft sign function |
|
Arctan function |
|
Nonzero sign log function |
|
Erf function |
|
Piecewise leaky relu function |
|
Squarewave Fourier series |
|
S2NN surrogate spiking function |
|
q-PseudoSpike surrogate function |
|
Leaky ReLU function |
|
Log-tailed ReLU function |
|
ReLU gradient function |
|
Gaussian gradient function |
|
Multi-Gaussian gradient function |
|
Inverse-square surrogate gradient |
|
Slayer surrogate gradient function |
|

