LoopOverTime#
- class brainpy.LoopOverTime(target, out_vars=None, no_state=False, t0=0.0, i0=0, dt=None, shared_arg=None, data_first_axis='T', name=None, jit=True, remat=False)[source]#
Transform a single step
DynamicalSysteminto a multiple-step forward propagationBrainPyObject.Note
This object transforms a
DynamicalSysteminto aBrainPyObject.If the target has a batching mode, before sending the data into the wrapped object, reset the state (
.reset_state(batch_size)) with the same batch size as in the given data.For more flexible customization, we recommend users to use
for_loop(), orDSRunner.Examples
This model can be used for network training:
>>> import brainpy as bp >>> import brainpy.math as bm >>> >>> n_time, n_batch, n_in = 30, 128, 100 >>> model = bp.Sequential(l1=bp.layers.RNNCell(n_in, 20), >>> l2=bm.relu, >>> l3=bp.layers.RNNCell(20, 2)) >>> over_time = bp.LoopOverTime(model, data_first_axis='T') >>> over_time.reset_state(n_batch) (30, 128, 2) >>> >>> hist_l3 = over_time(bm.random.rand(n_time, n_batch, n_in)) >>> print(hist_l3.shape) >>> >>> # monitor the "l1" layer state >>> over_time = bp.LoopOverTime(model, out_vars=model['l1'].state, data_first_axis='T') >>> over_time.reset_state(n_batch) >>> hist_l3, hist_l1 = over_time(bm.random.rand(n_time, n_batch, n_in)) >>> print(hist_l3.shape) (30, 128, 2) >>> print(hist_l1.shape) (30, 128, 20)
It is also able to used in brain simulation models:
>>> import brainpy as bp >>> import brainpy.math as bm >>> import matplotlib.pyplot as plt >>> >>> hh = bp.neurons.HH(1) >>> over_time = bp.LoopOverTime(hh, out_vars=hh.V) >>> >>> # running with a given duration >>> _, potentials = over_time(100.) >>> plt.plot(bm.as_numpy(potentials), label='with given duration') >>> >>> # running with the given inputs >>> _, potentials = over_time(bm.ones(1000) * 5) >>> plt.plot(bm.as_numpy(potentials), label='with given inputs') >>> plt.legend() >>> plt.show()
(
Source code,png,hires.png,pdf)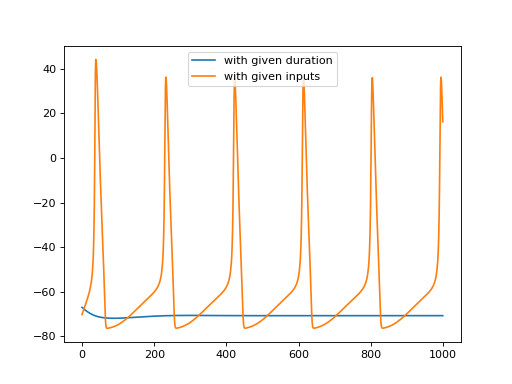
- Parameters:
target (DynamicalSystem) – The target to transform.
no_state (bool) –
Denoting whether the target has the shared argument or not.
For ANN layers which are no_state, like
DenseorConv2d, set no_state=True is high efficiently. This is because \(Y[t]\) only relies on \(X[t]\), and it is not necessary to calculate \(Y[t]\) step-bt-step. For this case, we reshape the input from shape = [T, N, *] to shape = [TN, *], send data to the object, and reshape output to shape = [T, N, *]. In this way, the calculation over different time is parralelized.
out_vars (PyTree) – The variables to monitor over the time loop.
t0 (float, optional) – The start time to run the system. If None,
twill be no longer generated in the loop.i0 (int, optional) – The start index to run the system. If None,
iwill be no longer generated in the loop.dt (float) – The time step.
shared_arg (dict) – The shared arguments across the nodes. For instance, shared_arg={‘fit’: False} for the prediction phase.
data_first_axis (str) – Denoting the type of the first axis of input data. If
'T', we treat the data as (time, …). If'B', we treat the data as (batch, time, …) when the target is in Batching mode. Default is'T'.name (str) – The transformed object name.
